The Chemistry of microbial dark matter
Marine sponges are rich sources of bioactive natural products
Most of these sponges are not available in quantities that permit drug development and production. We showed that symbiotic bacteria are the true producers of many of sponge-derived compounds. Since these symbionts usually do not grow in culture, we develop efficient methods
- to isolate biosynthetic gene clusters from sponge metagenomes (= multiple genomes),
- to identify the producers,
- and to express the genes in culturable bacteria that serve as renewable supplies.
These insights create new avenues for sustainable, biotechnological production of rare marine drug candidates - one of the lab's current priorities.
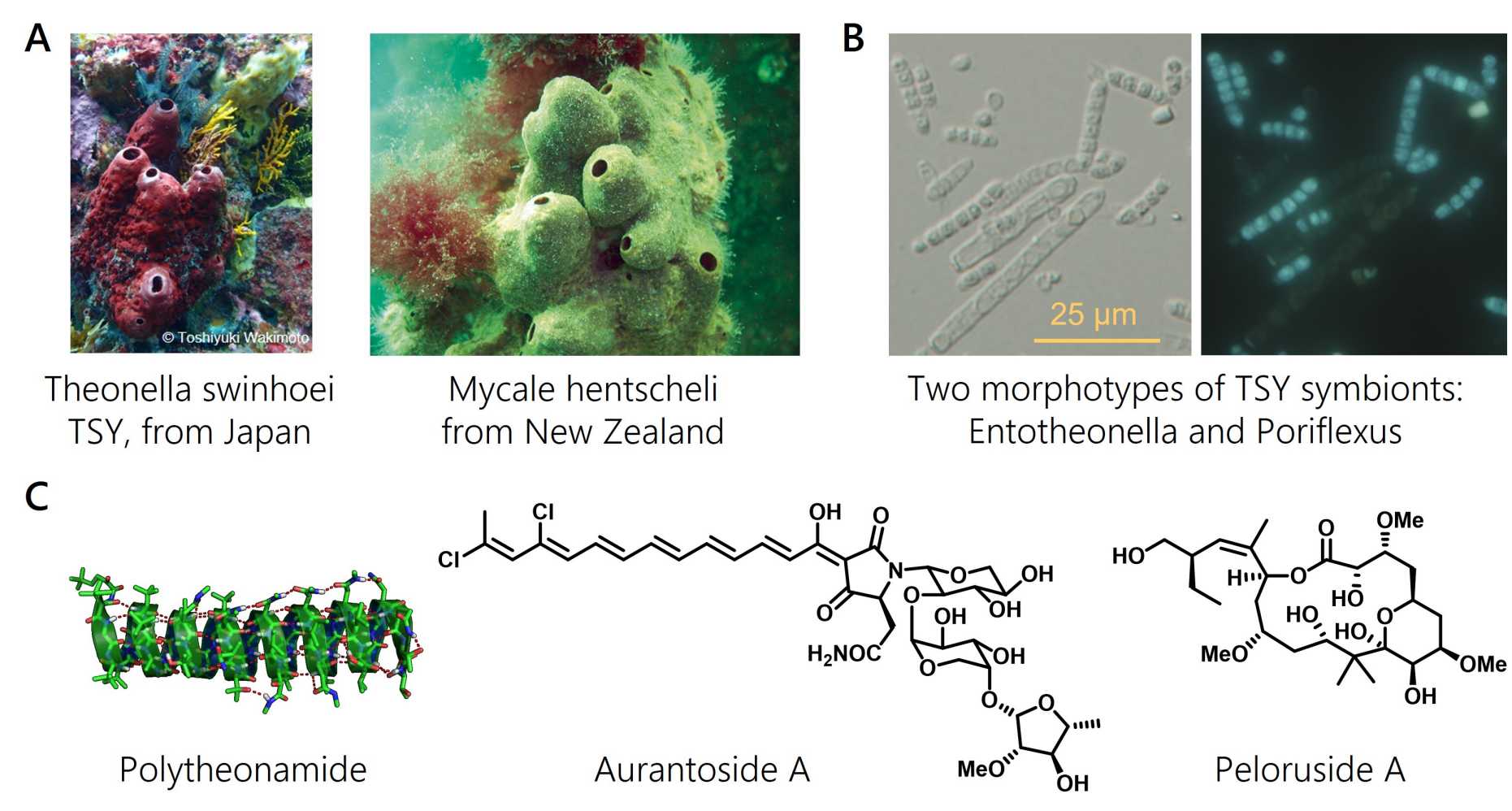
To date, our collaborative work has uncovered three distinct talented lineages: Entotheonella, Poriflexus, and Eudoremicrobiaceae. Entotheonella symbionts belong to a new, uncultivated candidate phylum that we have named 'Tectomicrobia'. Being present in many sponges, each Entotheonella variant produces distinct, large sets of bioactive compounds. We also showed that some chemically rich sponges, like Mycale hentscheli, associate with multiproducer microbiomes in which many members jointly synthesize a toxin cocktail.
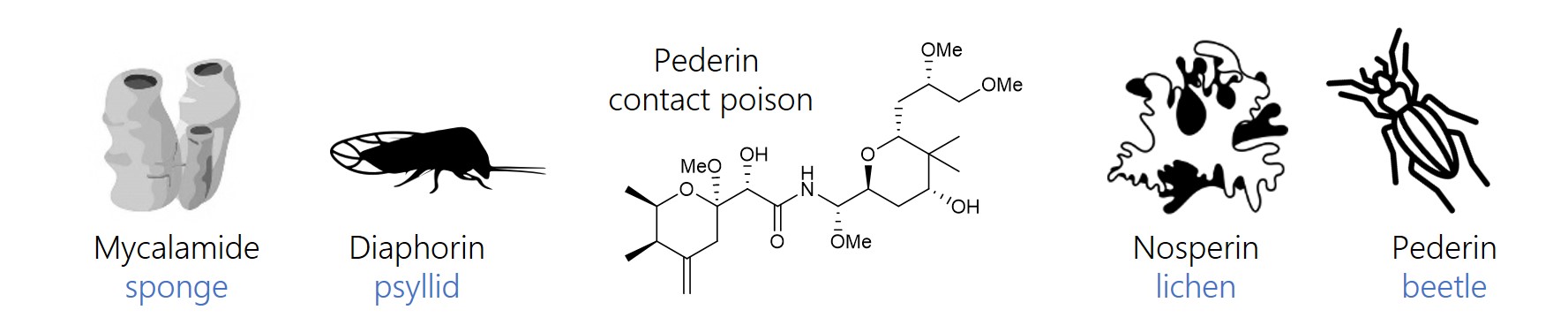
In addition to sponges, we contributed insights into metabolic functions of bacteria associated with various other hosts, including insects, lichens, and plants.
Related publications (selection)
Peters EE, Cahn JKB, Lotti A, Gavriilidou A, Steffens UAE, Loureiro C, Schorn MA, Cárdenas P, Vickneswaran N, Crews P, Sipkema D, Piel J. "Distribution and diversity of 'Tectomicrobia', a deep-branching uncultivated bacterial lineage harboring rich producers of bioactive metabolites." ISME Commun. (2023) 3, 1, 50. external pageAbstractcall_made
Kogawa M, Miyaoka R, Hemmerling F, Ando M, Yura K, Ide K, Nishikawa Y, Hosokawa M, Ise Y, Cahn JKB, Takada K, Matsunaga S, Mori T, Piel J, Takeyama H. "Single-cell metabolite detection and genomics reveals uncultivated talented producer." PNAS Nexus (2022) 1, 1, pgab007. external pageAbstractcall_made
Dieterich CL, Probst SI, Ueoka R, Sandu I, Schäfle D, Molin MD, Minas HA, Costa R, Oxenius A, Sander P, Piel J. "Aquimarins, Peptide Antibiotics with Amino-Modified C-Termini from a Sponge-Derived Aquimarina sp. Bacterium." Angew. Chem. Int. Ed. (2022) 61, 8, e202115802. external pageAbstractcall_made
Rust M, Helfrich EJN, Freeman MF, Nanudorn P, Field CM, Rückert C, Kündig T, Page MJ, Webb VL, Kalinowski J, Sunagawa S, Piel J. "A multiproducer microbiome generates chemical diversity in the marine sponge Mycale hentscheli." PNAS (2020) 117, 17, 9508-9518. external pageAbstractcall_made
Mori T, Cahn JKB, Wilson MC, Meoded RA, Wiebach V, Martinez AFC, Helfrich EJN, Albersmeier A, Wibberg D, Dätwyler S, Keren R, Lavy A, Rückert C, Ilan M, Kalinowski J, Matsunaga S, Takeyama H, Piel J. "Single-bacterial genomics validates rich and varied specialized metabolism of uncultivated Entotheonella sponge symbionts." PNAS (2018) 8, 1718-1723. external pageAbstractcall_made
Ueoka R, Uria AR, Reiter S, Mori T, Karbaum P, Peters EE, Helfrich EJ, Morinaka BI, Gugger M, Takeyama H, Matsunaga S, Piel J. "Metabolic and evolutionary origin of actin-binding polyketides from diverse organisms." Nat. Chem. Biol. (2015) 9, 705-12. external pageAbstractcall_made
M.C. Wilson, T. Mori, C. Rückert, A.R. Uria, M.J. Helf, K. Takada, C. Gernert, U.A.E. Steffens, N. Heycke, S. Schmitt, C. Rinke, E.J.N. Helfrich, A.O. Brachmann, C. Gurgui, T. Wakimoto, M. Kracht, M. Crüsemann, U. Hentschel, I. Abe, S. Matsunaga, J. Kalinowski, H. Takeyama, and J. Piel "An environmental bacterial taxon with a large and distinct metabolic repertoire". Nature (2014) 506, 7486, 58-62. external pageAbstractcall_made
M.F. Freeman, C. Gurgui, M.J. Helf, B.I. Morinaka, A.R. Uria, N.J. Oldham, H.-G. Sahl, S. Matsunaga, J. Piel “Metagenome mining reveals polytheonamides as modified ribosomal peptides” Science (2012) 338, 387-390. external pageAbstractcall_made
K.M. Fisch, C. Gurgui, N. Heycke, S.A. van der Sar, S.A. Anderson, V.L. Webb, S. Taudien, M. Platzer, B.K. Rubio, S.J. Robinson, P. Crews, J. Piel "Polyketide assembly lines of uncultivated sponge symbionts from structure-based gene targeting" Nat. Chem. Biol. (2009) 7, 494-501. external pageAbstractcall_made
Zimmermann K, Engeser M, Blunt JW, Munro MH, Piel J. "Pederin-type pathways of uncultivated bacterial symbionts: analysis of O-methyltransferases and generation of a biosynthetic hybrid." JACS (2009) 131, 8, 2780-1. external pageAbstractcall_made
J. Piel, D. Hui, G. Wen, D. Butzke, M. Platzer, N. Fusetani, S. Matsunaga "Antitumor polyketide biosynthesis by a bacterial symbiont of the marine sponge Theonella swinhoei” PNAS (2004) 101, 16222-16227. external pageAbstractcall_made
Ueoka R, Sondermann P, Leopold-Messer S, Liu Y, Suo R, Bhushan A, Vadakumchery L, Greczmiel U, Yashiroda Y, Kimura H, Nishimura S, Hoshikawa Y, Yoshida M, Oxenius A, Matsunaga S, Williamson RT, Carreira EM, Piel J. "Genome-based discovery and total synthesis of janustatins, potent cytotoxins from a plant-associated bacterium." Nat Chem. (2022), 10, 1193-1201. external pageAbstractcall_made
Ueoka R, Meoded RA, Gran-Scheuch A, Bhushan A, Fraaije MW, Piel J. "Genome Mining of Oxidation Modules in trans-Acyltransferase Polyketide Synthases Reveals a Culturable Source for Lobatamides." Angew. Chem. Int. Ed. (2020) 59, 20, 7761-7765. external pageAbstractcall_made
Ueoka R, Bhushan A, Probst SI, Bray WM, Lokey RS, Linington RG, Piel J. "Genome-Based Identification of a Plant-Associated Marine Bacterium as a Rich Natural Product Source." Angew. Chem. Int. Ed. (2018) 57, 44, 14519-14523. external pageAbstractcall_made
Helfrich EJN, Vogel CM, Ueoka R, Schäfer M, Ryffel F, Müller DB, Probst S, Kreuzer M, Piel J, Vorholt JA. "Bipartite interactions, antibiotic production and biosynthetic potential of the Arabidopsis leaf microbiome." Nat. Microbiol. (2018) 8, 909-919. external pageAbstractcall_made
Kampa A, Gagunashvili AN, Gulder TAM, Daolio C, Godejohann M, Miao VPW , Piel J, Andrésson ÓS "Metagenomic natural product discovery in lichen provides evidence for specialized biosynthetic pathways in diverse symbioses" Proc. Natl. Acad. Sci. U. S. A. (2013), 110, E3129-3137 external pageAbstractcall_made
Brachmann AO, Probst SI, Rüthi J, Dudko D, Bode HB, Piel J. "A Desaturase-Like Enzyme Catalyzes Oxazole Formation in Pseudomonas Indolyloxazole Alkaloids." Angew. Chem. Int. Ed. (2021) 60, 16, 8781-8785. external pageAbstractcall_made
A. Nakabachi, R. Ueoka, K. Oshima, R. Teta, A. Mangoni, M. Gurgui, N.J. Oldham, G. van Echten-Deckert, K. Okamura, K. Yamamoto, H. Inoue, M. Ohkuma, Y. Hongoh, S.-Y. Miyagishima, M. Hattori, J. Piel, T. Fukatsu "Defensive bacteriome symbiont with a drastically reduced genome" Curr. Biol. (2013) 23, 1478-1484. external pageAbstractcall_made
J. Piel "A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles" Proc Natl Acad Sci U S A. (2002) Oct 29;99(22):14002-7. external pageAbstractcall_made
